Evolution of a minimal cell
In this paper, Moger-Reischer build on previous research in synthetic biology where a minimal cell was constructed, and examine the impact of streamlining the genome on subsequent evolution of such minimal cells. Their results suggest that genome streamlining doesn’t prevent subsequent evolution of minimal cells.
Introduction
The complexity of living systems is related to genome size, and varies wildly between organisms. The simplest organisms—such as the ones studied by Moger-Reischer in this paper—possess the minimal number of genes for reproduction and survival in a given environment. One important implication of having such a minimal genome is that any mutation could lethally disrupt these functions.
The cell is the simplest independent unit of life, and previous research in synthetic biology has succeeded in constructing a viable minimal cell with a genome containing only the minimal number of genes required for survival. This accomplishment raised an important question: how does this streamlining of the genome affects an organism’s response to the forces of evolution? This is the question Moger-Reischer address in this paper.
The experiment
The experiment used by the authors was done on M. mycoides, a bacteria belonging to the Mollicutes. Specifically, the minimal cell used, called JCVI-syn3B and containing only 493 genes, is based on the non-minimal strain JCVI-syn1.0 containing 901 genes. This is the smallest genome of any organism that can be grown in a laboratory.
Two investigations were conducted to answer the following questions:
- whether genome streamlining—consisting of the removal of two DNA-replication genes, eight DNA repair genes and other genes of unknown function—altered the rate and spectrum of new mutations
- whether genome minimization altered the rate and mechanisms of evolution in response to natural selection, measured through whole-genome sequencing, estimates of population fitness and phenotypic changes in cell size.
First investigation
Mutation rate
The authors first conducted mutation accumulation experiments with populations of M. mycoides. They found that the number of mutations per nucleotide per generation for the minimal cell was indistinguishable from that of the non-minimal cell (Figure 1a). In other words, this means that the mutation rate was (surprisingly) not affected by genome minimization, which included the elimination of genes involved in replication fidelity.

Single-nucleotide mutations
While the composition of mutations such as insertions, deletions, and single-nucleotide mutations (SNMs) was not affected by genome minimization (Figure 1b), the various rates of SNMs themselves (which consisted of 88% of the total number of mutations) differed between the minimal and non-minimal cells (Figure 1c). Specifically, Moger-Reischer and colleagues found that for both cell types, mutations going from G or C to either A or T occurred at a higher rate than the ones going in the opposite direction, but that the magnitude of this bias was much greater in the minimal cell (100-fold bias) than in the non-minimal cell (30-fold bias).
Second investigation
Fitness
In the experiments conducted by the authors, the mutation rates were of \(3\times10^{-8}\) mutations per nucleotide per generation. Considering that the populations were of \(N=10^7\) individuals, this results in new mutations hitting every nucleotide in the genome more than 250 times during the 2000 generations of the evolution experiment. Put differently, neither cell type would thus be limited by the number of mutations per se, and differences in outcomes would be driven solely by alterations in genome content from the streamlining.
What they did next was to evolve the population for 2000 generations, then measure their fitness using two methods:
- by quantifying the maximum growth rate (\(\mu_{\text{max}}\)) every 65-130 generations, they could show that the minimization led to a 57% reduction in (\(\mu_{\text{max}}\))—but that both minimal and non-minimal cells subsequently increased their growth rate linearly in a similar fashion
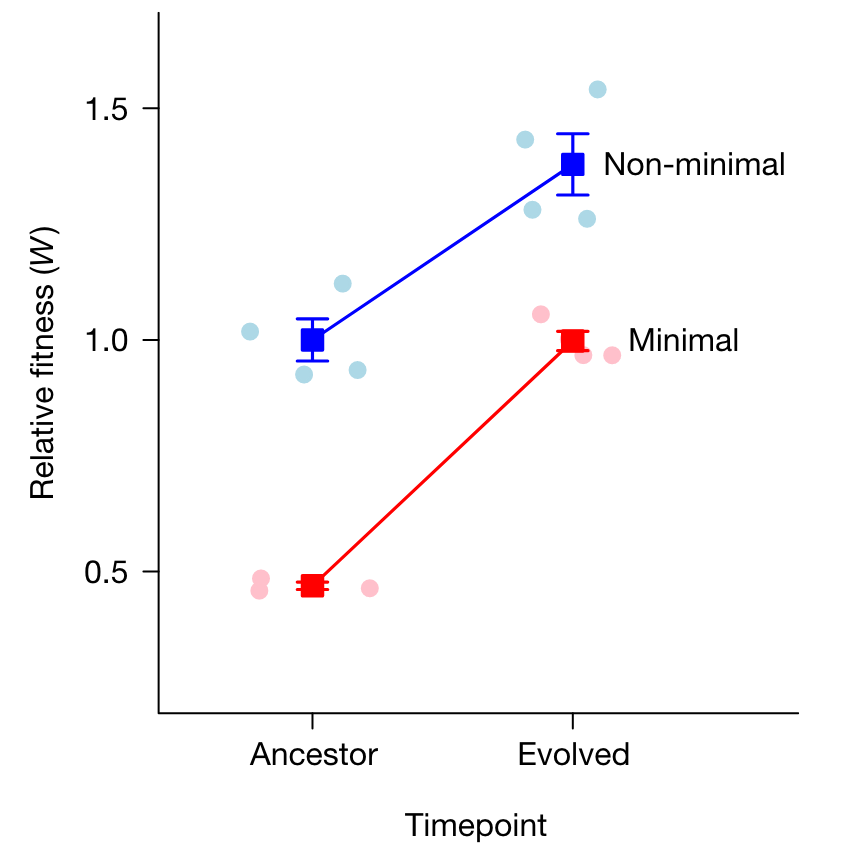
- by measuring relative fitness using head-to-head competition assays between the ancestal (generation 0) and evolved (generation 2000) populations, whereby they could similarly show a 53% decrease in fitness resulting from the minimization (Figure 2), after which the minimal cells adapted 39% more rapidly and recovered their fitness within 300 days.
These results tell an important story: they suggest that a streamlined genome is not inherently crippled and can perform as well as the non-minimalized cell after readaptation. Natural selection can thus quickly increase the fitness of even the simplest organisms.
Cell size
Moger-Reischer and colleagues also examined how cell size varied between minimal and non-minimal cells. We know that the size of single-celled organisms is variable, and linked to fitness in complex ways. For instance, in resource-rich environments it tends to be positively correlated with growth rate (an indicator of fitness). But this increase in cell-size, which accommodates more macromolecules needed for growth and division, also comes at a cost: it increases the surface-to-volume ratio, which reduces the efficiency of substrate diffusion.
In this case, genome streamlining was initially found to reduce cell diameter by 31%. After evolution, the size of non-minimal cells increased by 85%, while that of minimal cells did not appreciably change (Figure 3). The authors mention that one mutation, ftsZE315, was found to be responsible for 60% of evolved divergence in cell size. This would also indicate that the FtsZ protein (and ftsZ gene) has a central role in defining the cell size of M. mycoides. Cell size thus seems to evolve in a manner that is dependent on the genomic context.

Conclusion
The main message here is that natural selection during laboratory growth outweighed any deleterious effect of the genome disruption associated with streamlining. Moger-Reischer also mention that by combining evolution with synthetic biology we could improve gene characterization and the mapping of regulatory networks, which may ultimately be used to optimize living systems—for instance in the context of biotechnology.
Generalizing the results presented by the authors, we could also say that cellular functions are robust to streamlining over time, another feature which would be desirable in biotechnological/bioproduction. In other words, the results reported here suggest that genome streamlining does not constrain fitness, and does not preclude further diversification of populations over time.
Copyright: Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)
Author: Astrobiobites
Posted on: July 11, 2023